
Our new progress in understanding of the scallop byssus adhesion mechanism!
Recently, we published a research paper titled "Decoding the byssus fabrication by spatiotemporal secretome analysis of scallop foot" in Computational and Structural Biotechnology.
Secretome is involved in almost all physiological, developmental, and pathological processes, but to date there is still a lack of highly-efficient research strategy to comprehensively study the secretome of invertebrates. Adhesive secretion is a ubiquitous and essential physiological process in aquatic invertebrates with complicated protein components and unresolved adhesion mechanisms, making it a good subject for secretome profiling studies. Here we proposed a computational pipeline for systematic profiling of byssal secretome based on spatiotemporal transcriptomes of scallop. A total of 186 byssus-related proteins (BRPs) were identified, which represented the first characterized secretome of scallop byssal adhesion. Scallop byssal secretome covered almost all of the known structural elements and functional domains of aquatic adhesives, which suggested this secretome-profiling strategy had both high efficiency and accuracy. We revealed the main components of scallop byssus (including EGF-like domain containing proteins, the Tyr-rich proteins and 4C-repeats containing proteins) and the related modification enzymes primarily contributing to the rapid byssus assembly and adhesion. Spatiotemporal expression and co-expression network analyses of BRPs suggested a simultaneous secretion pattern of scallop byssal proteins across the entire region of foot and revealed their diverse functions on byssus secretion. In contrast to the previously proposed "root-initiated secretion and extension-based assembly” model, our findings supported a novel "foot-wide simultaneous secretion and in situ assembly” model of scallop byssus secretion and adhesion. Systematic analysis of scallop byssal secretome provides important clues for understanding the aquatic adhesive secretion process, as well as a common framework for studying the secretome of non-model invertebrates.
Prof. Shi Wang and Dr. Jing Wang are co-corresponding authors, and doctoral candidates Xiaoting Dai, Xuan Zhu and Dr. Lisui Bao are co-first authors. This research is supported by the Marine S&T Fund of Shandong Province for Pilot National Laboratory for Marine Science and Technology (Qingdao), National Natural Science Foundation of China, Key R&D Project of Shandong Province, China Agriculture Research System of MOF and MARA, and Taishan Scholar Project Fund of Shandong Province of China.
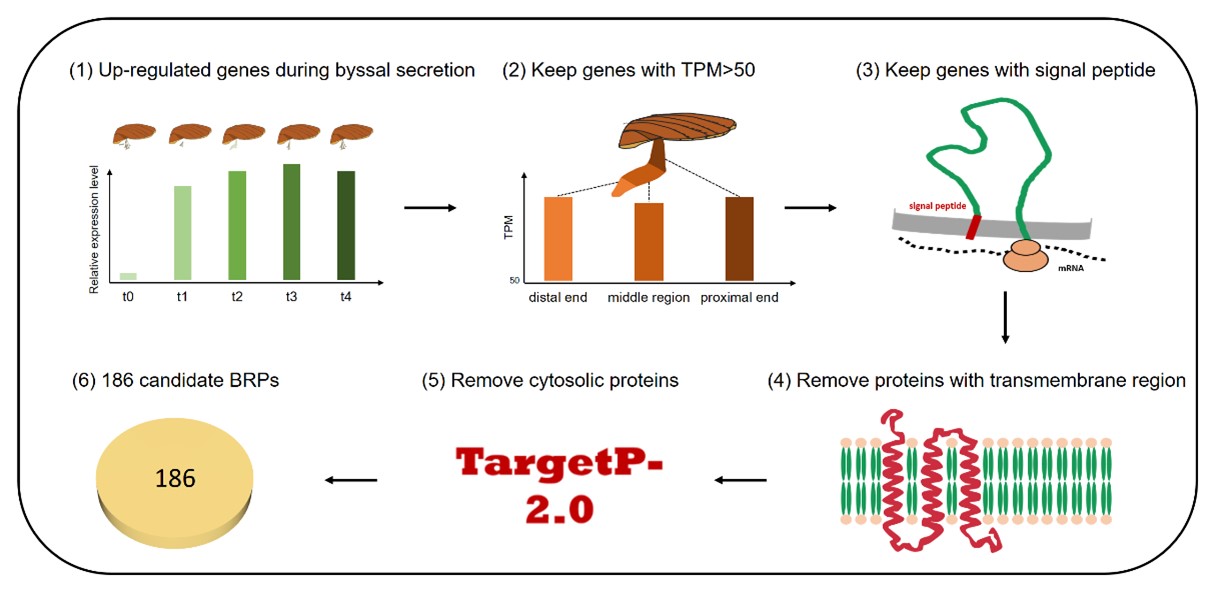
Figure 1. Profiling of byssal secretome based on spatiotemporal transcriptomes of scallop
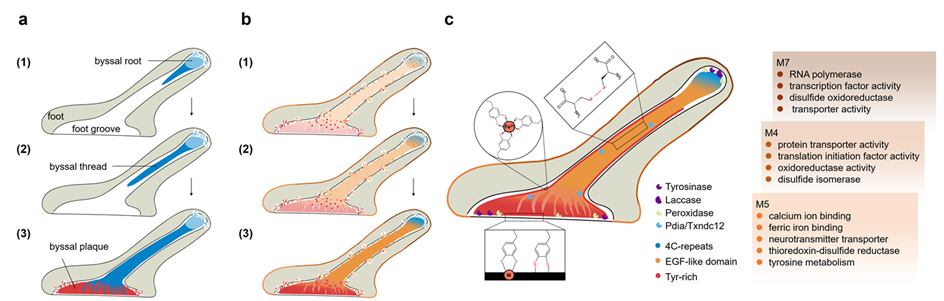
Figure 2. Model of scallop byssal secretion and adhesion
Dai X#, Zhu X#, Bao L#, Chen X, Miao Y, Li Y, Li Y, Lv J, Zhang L, Huang X, Bao Z, Wang S*, Wang J*.(2022) Decoding the byssus fabrication by spatiotemporal secretome analysis of scallop foot. Computational and Structural Biotechnology Journal. 20: 2713-2722.
https://www.sciencedirect.com/science/article/pii/S2001037022002045

