
Species-resolved sequencing of lowbiomass or degraded microbiomes using 2bRAD-M

On January 26, 2022, our team have developed a new metagenomics method (2bRAD-M) that can cost-effectively produce accurate, species-resolution, landscape-like taxonomic profiles for challenging microbiome samples. The method, published in the journal Genome Biology, overcome the bottlenecks of conventional metagenomic sequencing methods and greatly expands the opportunities in microbiome research in challenging settings. This work was in collaboration with Professor Jian Xu’s team at Qingdao Institute of BioEnergy and Bioprocess Technology, Chinese Academy of Sciences.
Metagenome sequencing, widely used to derive the taxonomic profile of microbiome, typically adopts two strategies that target (i) amplicons of phylogenetic “marker genes” (e.g., 16S/18S/ITS) or (ii) the whole genomes (whole metagenome shotgun; WMS). Although less costly, marker gene analyses can be limited in taxonomic resolution and susceptible to PCR bias; moreover, they are usually unable to capture a landscape-like view that includes bacteria, archaea, fungi, and virus. In contrast, by sequencing the total DNA, WMS can resolve species- or strain-level taxonomy and offers a landscape-like view that includes all domains of organisms. However, WMS is not efficient in tackling DNA samples that are low in biomass, heavily degraded, or dominated by host DNA. Furthermore, WMS is typically much more costly, due to the much higher sequencing volume required for covering the whole genome.
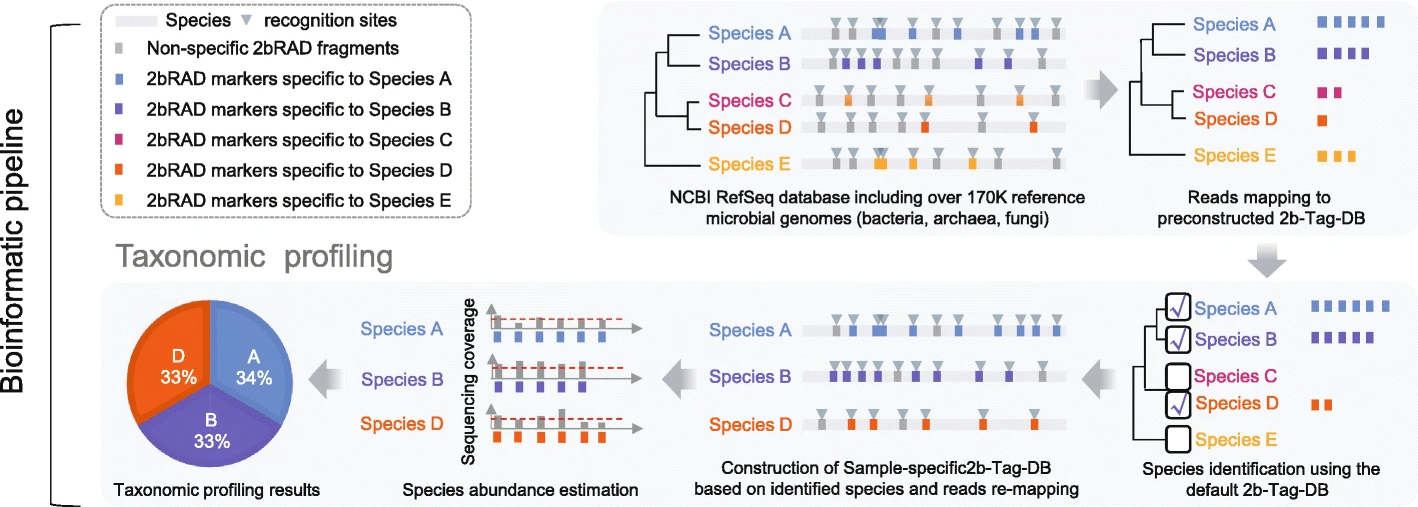
To address these challenges, here, our team developed a new metagenomics method (2bRAD-M), a highly reduced and cost-effective strategy which only sequences ~ 1% of metagenome and can simultaneously produce species-level bacterial, archaeal, and fungal profiles. 2bRAD-M can accurately generate species-level taxonomic profiles for otherwise hard-to-sequence samples with merely 1 pg of total DNA, high host DNA contamination, or severely fragmented DNA from degraded samples. Tests of 2bRAD-M on various stool, skin, environmental, and clinical FFPE samples suggest a successful reconstruction of comprehensive, high-resolution microbial profiles.
Professor Shi Wang (Sars-Fang Centre & MOE Key Laboratory of Marine Genetics and Breeding), Professor Jian Xu and Doctor Shi Huang (Single-Cell Center, CAS Key Laboratory of Biofuels and Shandong Key Laboratory of Energy Genetics, Qingdao Institute of BioEnergy and Bioprocess Technology, Chinese Academy of Sciences) are co-corresponding authors of this study. This work was funded by National Natural Science Foundation (NSFC), National Key Research and Development Program of China, Strategic Priority Research Program of the CAS, Sanya Yazhouwan Science and Technology City Management Foundation and Taishan Scholar Fund of Shandong Province of China.

