
We published new genome-wide profiling method of genetic and epigenetic variations: Multi-isoRAD
On October 6, 2016, our group have developed a technique that allows the preparation of five concatenated isoRAD tags for Illumina paired-end (PE) sequencing. The new technology, published in the journal Nature Protocols, addresses the issue of the original Multi-isoRAD protocol that it cannot be adapted for cost-effective PE sequencing and also offers researchers more power and flexibility in library configurations to meet specific research purposes. This is the third time that our team publish genomic technologies for non-model organisms, following the 2b-RAD published in Nature Methods in 2012 and MethylRAD published in Open Biology in 2015.
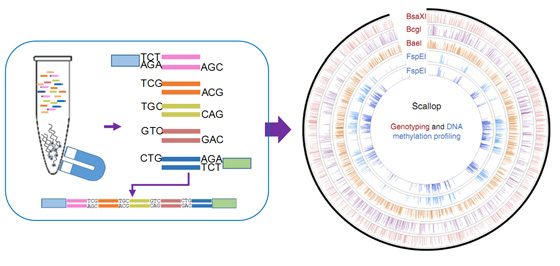
With the rapid development of high-throughput GBS methods during the past decade, the scientific community has witnessed huge success in numerous and diverse applications, addressing many scientific questions with unprecedented power and precision. The original Multi-isoRAD methodology (called 2b-RAD) was developed for genome-wide genotyping at minimal labor and cost. Just recently, our group has expanded its application to the epigenetics field and provided a GBS-based method (called MethylRAD) that allows quantification of DNA methylation levels. The isoRAD method is distinct from other GBS methods because of its use of special restriction enzymes to produce isolength tags, and sequencing of these uniform tags can bring many benefits. However, the relatively short tags produced by the original protocol are mostly suited to single-end (SE) sequencing (36–50 bp), and therefore they cannot efficiently match the gradually increased sequencing capacity of next-generation sequencing (NGS) platforms. To address this issue, we describe an advanced protocol named Multi-isoRAD that allows the preparation of five concatenated isoRAD tags for Illumina paired-end (PE) sequencing (100–150 bp). Multi-isoRAD inherits the advantages of isoRAD over many other GBS methods, such as almost all restriction sites being potentially assayable, lack of fragment size bias, tunable genome representation using selective adaptors, the ability to allow for extremely low DNA input (down to 1 ng) and low sensitivity to DNA degradation. With the new technical improvement of tag concatenation, Multi-isoRAD gains additional benefits, including the following: markedly reduced cost per tag, with a 20% reduction in the cost of library preparation and a 90% reduction in the cost of sequencing; extremely even read productivity across samples (multi-sample library); and multi-enzyme library preparation for increasing genome representation or simultaneously detecting genetic and epigenetic variations. Multi-isoRAD provides a cost-effective and highly-flexible method for genomics and molecular breeding research for non-model organisms.
Professor Zhenmin Bao and Professor Shi Wang are co-corresponding authors of this study. This research was granted by the National Natural Science Foundation of China, the National High Technology Research and Development Program of China 863 Program, the Natural Science Foundation for Distinguished Young Scholars of Shandong Province of China, and the AoShan Talents Program, supported by Qingdao National Laboratory for Marine Science and Technology.

